SNP Calendar / Timeline 
- E M96 49000 BCE
- E1 CTS9083 48500 BCE
- E1b P177 47200 BCE
- E1b1 P2 39900 BCE
- E1b1a V38 38000 BCE
- E1b1a1 M2 15500 BCE
- E1b1a1a M4901 13600 BCE
- E1b1a1a1 M180 8050 BCE
- E1b1a1a1a M4895 7380 BCE
- E1b1a1a1a1 M4706 7250 BCE
- E1b1a1a1a1c L485 7100 BCE
- E1b1a1a1a1c1 L514 6530 BCE
- E1b1a1a1a1c1? CTS7773 4460 BCE
- E1b1a1a1a1c1a1 M191 3780 BCE
- E1b1a1a1a1c1a1a U174 2770 BCE
- E1b1a1a1a1c1a1a3 CTS7305 2320 BCE
- E1b1a1a1a1c1a1a3a Z22455 2160 BCE
- E1b1a1a1a1c1a1a3a. SK1733 1690 CE L Cluster 0508
- E1b1a1a1a1c1a1a3a. Y131295 1750 CE
- E1b1a1a1a1c1a1a3a. SK1733 1690 CE L Cluster 0508
- E1b1a1a1a1c1a1a3a Z22455 2160 BCE
- E1b1a1a1a1c1a1a3 CTS7305 2320 BCE
- E1b1a1a1a1c1a1a U174 2770 BCE
- E1b1a1a1a1c1a1 M191 3780 BCE
- E1b1a1a1a1c1? CTS7773 4460 BCE
- E1b1a1a1a1c1 L514 6530 BCE
- E1b1a1a1a1c L485 7100 BCE
- E1b1a1a1a2 CTS229 6860 BCE
- E1b1a1a1a2a U175 6720 BCE
- E1b1a1a1a2a1 P277 4340 BCE
- E1b1a1a1a2a1a M4232 4060 BCE
- E1b1a1a1a2a1a3 M4257 3940 BCE
- E1b1a1a1a2a1a3b M4041 3850 BCE
- E1b1a1a1a2a1a3b1 CTS2504 2610 BCE
- E1b1a1a1a2a1a3b1a U290 2500 BCE
- E1b1a1a1a2a1a3b1d Z1788 2320 BCE
- E1b1a1a1a2a1a3b1d1 ?
- E1b1a1a1a2a1a3b1d1c CTS11328 1960 BCE
- E1b1a1a1a2a1a3b1d1c1 CTS8026 1150 BCE
- E1b1a1a1a2a1a3b1d1c1a CTS10652 840 BCE L Cluster 0510
- E1b1a1a1a2a1a3b1d1c1 CTS8026 1150 BCE
- E1b1a1a1a2a1a3b1d1c CTS11328 1960 BCE
- E1b1a1a1a2a1a3b1d1 ?
- E1b1a1a1a2a1a3b1 CTS2504 2610 BCE
- E1b1a1a1a2a1a3b M4041 3850 BCE
- E1b1a1a1a2a1a3 M4257 3940 BCE
- E1b1a1a1a2a1a M4232 4060 BCE
- E1b1a1a1a2a1 P277 4340 BCE
- E1b1a1a1a2a U175 6720 BCE
- E1b1a1a1a1 M4706 7250 BCE
- E1b1a1a1a M4895 7380 BCE
- E1b1a1a1 M180 8050 BCE
- E1b1a1a M4901 13600 BCE
- E1b1a1 M2 15500 BCE
- E1b1b M215 32800 BCE
- E1b1b1 M35 22700 BCE
- E1b1b1a? V68 18600 BCE
- E1b1b1a1 M78 12300 BCE
- E1b1b1a1b Z1919 10800 BCE
- E1b1b1a1b1 L618 7290 BCE
- E1b1b1a1b1a V13 3140 BCE
- E1b1b1a1b1a. CTS8814 2920 BCE
- E1b1b1a1b1a.. CTS5856 2410 BCE L??? Cluster 0504 do more testing
- E1b1b1a1b1a. CTS8814 2920 BCE
- E1b1b1a1b1a V13 3140 BCE
- E1b1b1a1b1 L618 7290 BCE
- E1b1b1a1b Z1919 10800 BCE
- E1b1b1a1 M78 12300 BCE
- E1b1b1a? V68 18600 BCE
- E1b1b1 M35 22700 BCE
- E1b1a V38 38000 BCE
- E1b1 P2 39900 BCE
- E1b P177 47200 BCE
- E1 CTS9083 48500 BCE
The SNP calendar to the right shows the relationship between ISOGG long forms, E SNPs, and TMRCA estimates. SNP analysis is in a constant state of transition, so you will see differences between ISOGG, YFull, and FTDNA data. TMRCA estimates continue to be recalculated, depending on the amount of data YFull has available to analyze. Not all SNPs starting from E are shown, and there are enormous time gaps between some TMRCA estimates. Terminal SNPs that are the signature of project lineages are marked with L.
Haplogroup E project members constitute roughly 8% of Collins project members overall. The project has identified three divisions:
• Members with terminal SNP E-SK1733 or are close STR matches form a LINEAGE and constitute the largest project E haplogroup cluster. Some members have shared geographic origins in the southeastern United States, particularly northeastern counties of Tennessee.
• Members under SNP E-CTS10652 form a second, smaller LINEAGE. Members have shared an origin of Morgan County, Kentucky.
• Remaining haplogroup E members are UNMATCHED and have not done much further Y testing or shared much further information.
Haplogroup E project members can greatly expand the data on this page through Expert Level Participation (see #5).
Modal for CTS5856
This is project Cluster 0504. The project does not have active participants who have tested Y111 or more STR markers. Until this changes, only the cluster modal is shown.
| Table #1 Haplotypes E Modal and CTS10652 Collins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 13 | 24 | 13 | 10 | 16-17 | 11 | 12 | 12 | 12 | 11 | 30 | 15 | 9-9 | 11 | 11 | 26 | 14 | 20 | 31 | 14-16-16-17 | 9 | 11 | 19-21 | 16 | 12 | 17 | 20 | 32-35 | 11 | 10 | 10 | 8 | 15-15 | 8 | 11 | 10 | 8 | 12 | 10 | 22-23 | 17 | 11 | 12 | 12 | 17 | 7 | 12 | 22 | 18 | 13 | 13 | 12 | 14 | 11 | 11 | 11 | 11 | 34 | 15 | 8 | 15 | 11 | 24 | 27 | 19 | 13 | 12 | 12 | 11 | 12 | 9 | 13 | 11 | 10 | 11 | 12 | 31 | 10 | 13 | 18 | 14 | 11 | 10 | 19 | 16 | 21 | 12 | 24 | 13 | 14 | 16 | 24 | 14 | 23 | 18 | 12 | 14 | 17 | 9 | 12 | 11 | |||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Modal for SK1733 
This is project Cluster 0508. The table below shows the 111 marker modal for the E-SK1733 lineage.
This cluster is believed to be linked to the Melungeons. Members of this cluster are encouraged to join the Melungeon project.
| Table #1 Haplotypes E Modal SK1733 Collins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 15 | 20 | 17 | 10 | 15-17 | 11 | 12 | 12 | 14 | 11 | 31 | 16 | 8-10 | 11 | 11 | 26 | 14 | 21 | 32 | 14-16-17-18 | 11 | 10 | 19-21 | 16 | 13 | 16 | 17 | 33-33 | 11 | 11 | 10 | 8 | 16-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 20-22 | 20 | 11 | 12 | 13 | 16 | 7 | 12 | 24 | 23 | 14 | 12 | 12 | 13 | 9 | 11 | 11 | 12 | 33 | 14 | 8 | 15 | 12 | 24 | 27 | 20 | 13 | 12 | 11 | 12 | 11 | 8 | 11 | 11 | 10 | 11 | 12 | 30 | 9 | 11 | 20 | 14 | 10 | 10 | 22 | 15 | 18 | 12 | 23 | 13 | 12 | 15 | 24 | 14 | 22 | 18 | 13 | 15 | 19 | 9 | 13 | 11 | ||||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
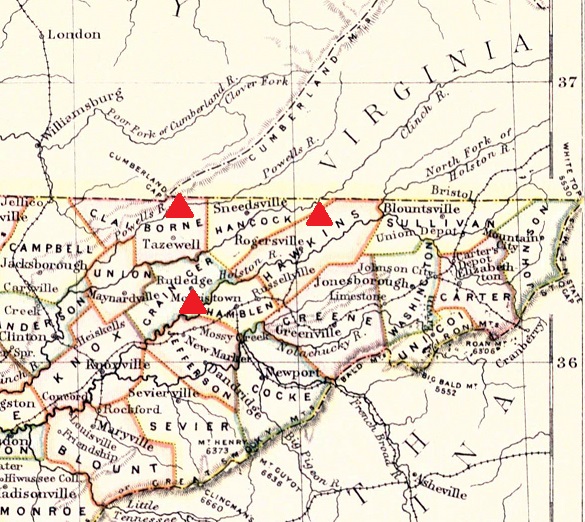 |
The map shows the ancestral origins of some of our E-SK1733 project members in northeastern Tennessee. Haplogroup E members of the Collins project can improve the data on this page by testing more markers and becoming Expert Level Participants. |
Haplotype Table for CTS10652 
This is project Cluster 0510. Our information on Collinses in this haplogroup is quite limited. There is one pedigree in the project. The haplotype table shows one Expert Level participant compared against a modal haplotype for the cluster. There is a Genetic Distance of 2 from the modal.
| Table #1 Haplotypes E Modal and CTS10652 Collins | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| relative position | 1 | 2 | 3 | 4 | 5-6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14-15 | 16 | 17 | 18 | 19 | 20 | 21 | 22-25 | 26 | 27 | 28-29 | 30 | 31 | 32 | 33 | 34-35 | 36 | 37 | 38 | 39 | 40-41 | 42 | 43 | 44 | 45 | 46 | 47 | 48 | 49-50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | 63 | 64 | 65 | 66 | 67 | 68 | 69 | 70 | 71 | 72 | 73 | 74 | 75 | 76 | 77 | 78 | 79 | 80 | 81 | 82 | 83 | 84 | 85 | 86 | 87 | 88 | 89 | 90 | 91 | 92 | 93 | 94 | 95 | 96 | 97 | 98 | 99 | 100 | 101 | 102 | 103 | 104 | 105 | 106 | 107 | 108 | 109 | 110 | 111 | relative position | |||||||||
| Short Tandem Repeat Markers (STRS) | DYS393 | DYS390 | DYS19 | DYS391 | DYS385 | DYS426 | DYS388 | DYS439 | DYS389I | DYS392 | DYS389II | DYS458 | DYS459 | DYS455 | DYS454 | DYS447 | DYS437 | DYS448 | DYS449 | DYS464 | DYS460 | Y-GATA-H4 | YCAII | DYS456 | DYS607 | DYS576 | DYS570 | CDY | DYS442 | DYS438 | DYS531 | DYS578 | DYF395S1 | DYS590 | DYS537 | DYS641 | DYS472 | DYF406S1 | DYS511 | DYS425 | DYS413 | DYS557 | DYS594 | DYS436 | DYS490 | DYS534 | DYS450 | DYS444 | DYS481 | DYS520 | DYS446 | DYS617 | DYS568 | DYS487 | DYS572 | DYS640 | DYS492 | DYS565 | DYS710 | DYS485 | DYS632 | DYS495 | DYS540 | DYS714 | DYS716 | DYS717 | DYS505 | DYS556 | DYS549 | DYS589 | DYS522 | DYS494 | DYS533 | DYS636 | DYS575 | DYS638 | DYS462 | DYS452 | DYS445 | Y-GATA-A10 | DYS463 | DYS441 | Y-GGAAT-1B07 | DYS525 | DYS712 | DYS593 | DYS650 | DYS532 | DYS715 | DYS504 | DYS513 | DYS561 | DYS552 | DYS726 | DYS635 | DYS587 | DYS643 | DYS497 | DYS510 | DYS434 | DYS461 | DYS435 | Short Tandem Repeat Markers (STRS) | |||||||||
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MODAL | 13 | 21 | 15 | 10 | 16-20 | 11 | 12 | 11 | 13 | 11 | 31 | 18 | 8-9 | 11 | 11 | 26 | 14 | 21 | 28 | 13-16-17-19 | 11 | 11 | 19-19 | 15 | 13 | 16 | 19 | 33-36 | 11 | 11 | 11 | 8 | 16-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 20-22 | 18 | 11 | 12 | 13 | 17 | 7 | 14 | 28 | 21 | 13 | 12 | 12 | 13 | 9 | 11 | 11 | 12 | 34 | 14 | 8 | 15 | 12 | 24 | 28 | 19 | 13 | 12 | 11 | 12 | 12 | 8 | 11 | 11 | 10 | 11 | 12 | 31 | 12 | 12 | 20 | 14 | 10 | 10 | 18 | 15 | 18 | 12 | 23 | 13 | 13 | 15 | 24 | 14 | 21 | 18 | 15 | 14 | 17 | 9 | 12 | 11 | MODAL | |||||||||
| 75577116 | 13 | 21 | 15 | 10 | 16-20 | 11 | 12 | 11 | 13 | 11 | 31 | 18 | 8-9 | 11 | 11 | 26 | 14 | 21 | 28 | 13-16-17-19 | 11 | 11 | 19-19 | 15 | 13 | 16 | 19 | 33-36 | 11 | 11 | 11 | 8 | 16-16 | 8 | 10 | 10 | 8 | 10 | 10 | 12 | 20-22 | 18 | 11 | 12 | 13 | 17 | 7 | 14 | 28 | 21 | 13 | 12 | 12 | 13 | 9 | 11 | 11 | 12 | 33 | 14 | 8 | 15 | 12 | 24 | 28 | 19 | 13 | 12 | 11 | 12 | 12 | 8 | 11 | 11 | 10 | 11 | 12 | 31 | 12 | 12 | 20 | 14 | 10 | 10 | 18 | 15 | 18 | 12 | 23 | 13 | 13 | 15 | 24 | 14 | 21 | 18 | 14 | 14 | 17 | 9 | 12 | 11 | 75577116 | |||||||||
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Collins at FTDNA | DNA Portal | Pedigrees | |
| See ABOUT for website information. | |||
| Copyright © 2015 - 2025 collins.dnagen.org. All rights reserved. | |||